
A Comprehensive Case-Study of GraphSage using PyTorchGeometric and Open-Graph-Benchmark
Estimated reading time: 15 minute
This blog post provides a comprehensive study on the theoretical and practical understanding of GraphSage, this notebook will cover:
- What is GraphSage
- Neighbourhood Sampling
- Getting Hands-on Experience with GraphSage and PyTorch Geometric Library
- Open-Graph-Benchmark’s Amazon Product Recommendation Dataset
- Creating and Saving a model
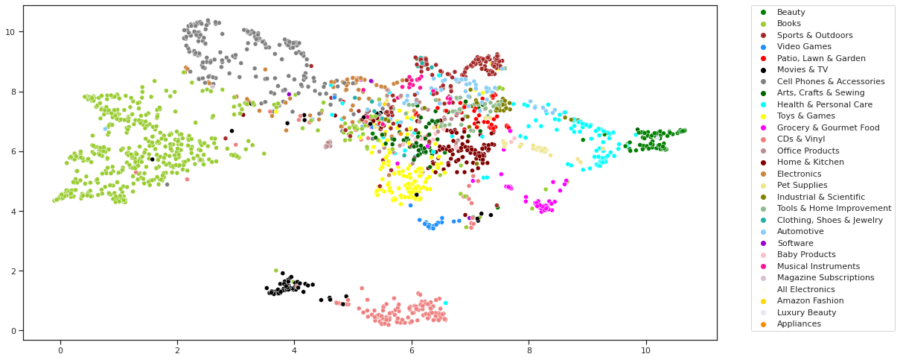
- Generating Graph Embeddings Visualizations and Observations
Hear More from the Author
Continue Reading
Inside the Avocado Grove: From Canada to Germany and the Digital Marketing of Avocados
Get the latest tutorials, blog posts and news:
